ABSTRACT
We describe
ELVIS(the Ecosystem
Location Visualization and Information System), a suite of
tools for constructing food webs for a given location. We
express both ELVIS input and output data in OWL, thereby
enabling its integration with other semantic web resources. In
particular, we describe using a Triple Shop application to
answer SPARQL queries from a collection of semantic web
documents. This is an end-to-end case study offor the semantic webÕs utility for
ecological and environmental research.
Categories and Subject Descriptors
H. Information Systems
H.4 Information System Applications
H.4.m miscellaneous
General Terms
Algorithms, Design, Experimentation, Human Factors, Standardization,
Keywords
Food webs - Ecological forecasting - Semantic web - Ontologies - Invasive Species - Biodiversity - Service Oriented Design
1. INTRODUCTION
SPIRE
(Semantic Prototypes in
Research Ecoinformatics)-http://spire.umbc.edu) is a
distributed, interdisciplinary research project tasked with
building semantic web prototypes for invasive species
science. {SPIRE: Semantic Prototypes in Researchh
Ecoinformatics 2006 #6010}
.
Our main integrating suite of tools is ELVIS (the Ecosystem Location Visualization Information System). ELVIS is motivated by the belief that food web structure plays a role in the success or failure of potential species invasions. Because very few ecosystems have been the subject of empirical food web studies, response teams are typically unable to get quick answers to questions like Òwhat are likely prey and predator species of the invader in the new environment?Ó
The core data has been
integrated from publicly available sources and is now available
on the semantic web. We have constructed a platform for
investigating multiple algorithms for food web prediction.
Further, by exposing item-level data through several rich sets
of ecological and evolutionary ontologies, and by providing
these tools as web services, we enable integration with other
semantic web/web 2.0 applications, such as SwoogleWOOGLE. WE and FieldMarking
{Parr #6020}
.
We have developed a Òshopping
cartÓ application, Triple Shop, which allows a user to select
semantic web documents, and to issue SPARQL queries over their
union. Thus, we are able to integrate diverse ecoinformatics
data in response to ad-hoc queries.
1.1 Related Work
Previous work on data
integration in ecological informatics includes online data
repositories[2] and workflow [4] ontologies. Metadata allows
only the discovery of possibly interesting datasets and does
not provide the means to harvest the data itself. Individual
food web researchers maintain and share their own digital data
archives, in individualized data formats, though more
accessible standardized archives are beginning to
emerge[1]. There are
goodexcellent databases on invasive
species
{Invasive Species Specialist Group 2006
#6060}
]s (e.g. http://www.issg.org/database/welcome/)
but they areit is not automatically integrated with
information about non-invasive specieswith which they
interacywith
which they may interact;, nor is there web-based support for
modeling an invasive species, anywhere.
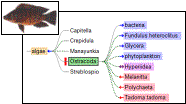
2. ELVIS
The task of providing food web information
for a user-specified location breaks into two distinct
problems: constructing a species list for a given location; and
constructing a food web from a given species list (and habitat
information).
 |
||||
|
||||
![]()
2.1 Species List Constructor
Our goal is to allow a user to input a location, and get back a species list for that location. This is a hard problem, typically ad-hoc, and relying on expert knowledge. There are, in general, three kinds of information that can be used to generate a species list: (i) park inventories; (ii) point locations, e.g. from specimen descriptions in museums and herbariums; and (iii) distribution maps generated by applying statistical techniques to point locations. We are integrating all of the above for California, and expect that the ontologies and synthesis strategies we have developed will apply to other states, and enable ELVIS to quickly spread beyond California.
2.2 Food Web Constructor
The Food Web Constructor (FWC) uses empirically known food web links to predict food web links not yet recorded.
|
Copyright is held by IW3C2. WWW 2006, May 22-26, 2006, Edinburgh, UK.
|
A user can choose which food web
studies to use for prediction or exclude from 257 datasets we
compiled from previously digitized literature.
{Cohen 1989 #6030}
,
{Vazquez 2005 #6040}
,{Dunne, Williams, et al. #6050}
.
T
Taaxa can be entered several
different ways: simple text lists, XML files, or food web
number. In this latter case we seek to reconstruct feeding
links based on the rest of the database and can therefore
assess the success rate of the different algorithms or model
parameters.
Each suspected link is reported, together with references to supporting evidence. Summary statistics of the food web are also reported.
2.3 Evidence Provider

Figure 2 shows the evidence for a predicted trophic link - namely, the actual link that was observed, the study in which it was published, and the relationship between the species in the observed link and the predicted link.
2.4 Technical Approach
The Species
List Constructor interacts with web services that provide a
variety of species informatione top half of ELVIS uses ontologies
(how?). OurThe current Food Web
Constructor algorithm uses taxonomic distances to weight
evidence supporting or failing to support links between
organisms. All data input, output, and taxonomies for Food Web
Constructor and Evidence Provider are available in OWL on
demand. ,
but at present the Ccalculations are
performed act on data residing in MySQL
databases. Scripts generate OWL documents from Animal Diversity
Web
{Myers 2003 #5080}
(http://www.animaldiversityweb.org)
via the
ETHAN ontology. The triple
shop is currently implemented using the
joseki(http://www.joseki.org/).
.Add Joseki web service and
www.sparqler.org?
3. Swoogle and Triple Shop
Swoogle (Google for the Semantic Web) is
ourthe semantic web search engine
[3](Swoogle ref). It allows users
to search for both ontologies and instance data (collectively
referred to as semantic web documents) along a variety of
parameters. Once documents are returned, a user can select
certain of them for inclusion into her ÒTriple ShopÓ, a sort of
shopping cart for RDF triples. We have built a stand-alone
version of the Triple Shop, which allows a user to specify the
URLs of arbitrary semantic web documents, and to issue SPARQL
queries against the union of those documents.
3.1 Using the Triple Shop to Integrate Food Web and Natural History Data
ELVIS illustrates the potential of the semantic web to support rapid querying of distributed scientific databases for a variety of scenarios. For example (Figure 3): Determine known predator-prey relationships among an invader and a specific group of native species in a particular habitat, as reported in previous studies.
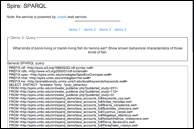
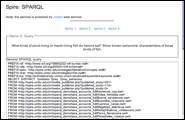
Figure 3. SPARQL query on ETHAN and SpireEcoConcepts OWL documents.
4. ACKNOWLEDGMENTS
This research was
supported by NSF ITRgrant
#0326460 and
matching funds received from the USGS
Nat.ional Bio.logical Information
Infrastructure.
1.
REFERENCES
[1]
Dunne, J. A. The network
structure of food webs. In: Ecological
Networks: Linking Structure to Dynamics in Food Webs, eds.
Pascual, M. and Dunne, J. A. Oxford
University Press, 2005.pp. 27-86.
[2]
Jones , Matthew
.B.; Berkley,
C.had;
Bojilova, J.Jivka;
Schildhauer,
M. ark
P. 2001. Managing scientific
metadata. IEEE Internet Computing. Vol: 5(5).
Pages 59-68.
[1]
Li Ding et
al., "Swoogle: A Search and Metadata Engine
for the Semantic Web", Proceedings of the Thirteenth ACM
Conference on Information and Knowledge Management, November
2004.
[3]
[4]Ludaescher, B. et al. 2004.
Scientific workflow management and the Kepler system.
Concurrency and Computation: Practice and
Experience.
Ludaescher,
B. Bertram; Altintas, Ilkay;
Berkley, Chad; Higgins, Dan; Jaeger, Efrat; Jones, Matthew B.;
Lee, Edward A.; Tao, Jing; Zhao,
Yanget
al.In press
2004. Scientific workflow
management and the Kepler system. Concurrency and Computation:
Practice and
Experience.Anderson, R.E. Social impacts of computing: Codes of
professional ethics. Social Science Computing Review, 2 (Winter
1992),
453-469.
ACM SIG
PROCEEDINGS template.
http://www.acm.org/sigs/pubs/proceed/template.html.
Conger., S., and Loch, K.D. (eds.). Ethics and computer use.
Commun. ACM 38, 12 (entire
issue).
Mackay,
W.E. Ethics, lies and videotape... in Proceedings of CHI
'95 (Denver CO, May 1995), ACM Press,
138-145.
Schwartz, M., and Task Force on Bias-Free Language.
Guidelines for Bias-Free Writing. Indiana University Press,
Bloomington, IN, 1995.